IntOMICS: A Bayesian Framework for Reconstructing Regulatory Networks Using Multi-Omics Data
 Concept
Concept
Abstract
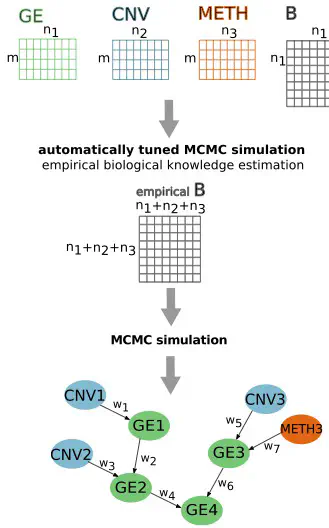
Integration of multi-omics data can provide a more complex view of the biological system consisting of different interconnected molecular components. We present a new comprehensive R/Bioconductor-package, IntOMICS, which implements a Bayesian framework for multi-omics data integration. IntOMICS adopts a Markov Chain Monte Carlo sampling scheme to systematically analyze gene expression, copy number variation, DNA methylation, and biological prior knowledge to infer regulatory networks. The unique feature of IntOMICS is an empirical biological knowledge estimation from the available experimental data, which complements the missing biological prior knowledge. IntOMICS has the potential to be a powerful resource for exploratory systems biology.