Molecular portraits of major morphological components of colon tumors
Funding: GACR (GA19-08646S); Principal investigator: Vlad Popovici
Partners: RECETOX - MMCI - CEITEC
Data available here or directly here
Background
Solid tumor manifest various degrees of heterogeneity and several taxonomies have been proposed for trying to group the tumors into more homogeneous and comprehenisible categories. Colorectal cancer (CRC) is not different. In fact, due to its usually slower progression and somehow later detection, it acquires more mutations and manifests higher intra-tumor morphological diversity then some other cancers.
Project
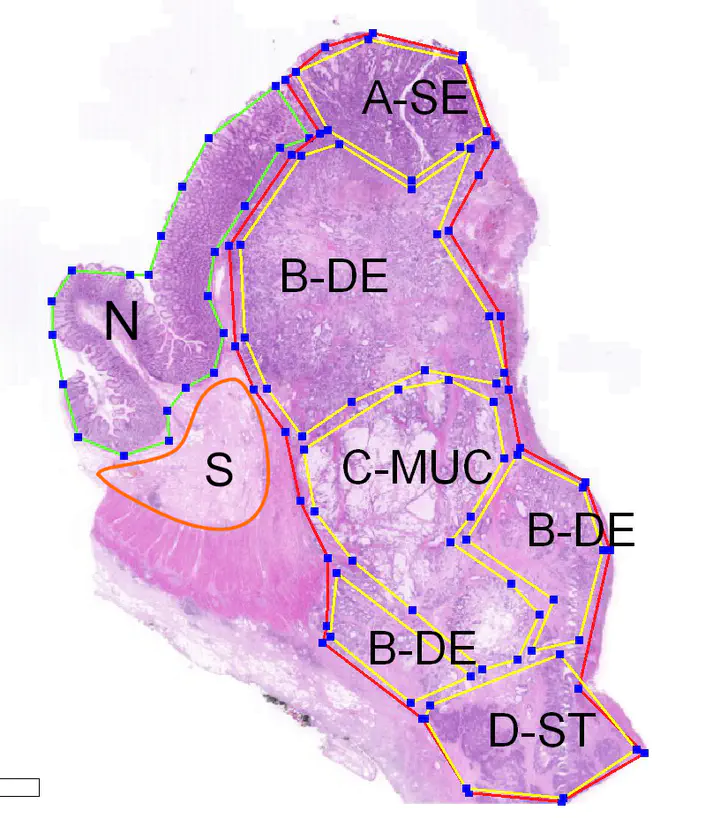
In this project, we investigate the CRC heterogeneity starting with its morphological features. Using the morphology as an anchor for tumor sampling, we explore the diversity of the molecular programs that drive the tumor evolution. We have obtained about 200 expression profiles from over 100 different tumors, representing six different morphotypes (complex tubular, desmoplastic, mucinous, papillary, serrated and solid/trabecular). In addition, we have also profiles tumor-adjacent normal tisse and tumor-associated stroma for building reference profiles.
Main results
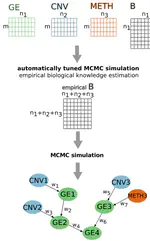
The morphology-guided tumor sampling allows for a finer resolution than whole-tumor profiling. On the other hand, the single-cell assays are able to extract very specific profiles from tiny tumor regions however at the cost of losing much of the phenotypical context. The bioinformatics analyses revealed a high degree of variability in gene expression between regions. More importantly, even within the same tumor (same section), we were able to identify differences in major pathways (e.g. MAPK, WNT, TGFb). Since these pathways are fundamental in tumor classification (e.g. by consensus molecular subtypes, or by prognostic signatures), it comes with no suprise that these classifiers vary also across regions, even within the tumor. These results call for a deeper investigation of the impact of intra-tumoral heterogeneity on the molecular-/expression-based decision systems.