Clinical implications of intra-tumour heterogeneity in colon cancer
Funding: AZV (NV19-03-00298); Principal investigator: Vlad Popovici
Partners: RECETOX - MMCI - CEITEC
Background
Differences that exist both between and within tumours of the same cancer type are a major hurdle towards proper patient selection for a given treatment and for developing more targeted therapies. Depending on the perspective under which these differences are investigated, different categorisation paradigms have emerged. The systematisation of clinical and histopathological parameters led to the definition of the current TNM staging system which presently constitutes the gold standard for diagnosis and prognosis. At a different scale, the intra-tumour heterogeneity (ITH) has been a long-time recognised phenomenon, but its tremendous impact on diagnosis and treatment outcome only recently started to be systematically investigated. Experiments such as single-cell or surgical tumour multi-sampling sequencing portrayed a highly heterogeneous picture of the various cancers with trunk and branch driver mutations distributed non-uniformly in time and space. In colon cancer, while KRAS, NRAS and BRAF appear to be trunk driver mutations, TP53 and PIK3CA seem to be branch driver mutations.
Project
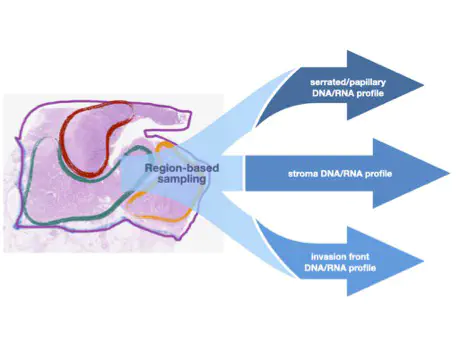
The overarching objective of this project is to explore and characterise the impact of ITH on the clinical decisions regarding prognosis (for stage II patients) and personalised treatment (for metastatic patients). Such analyses require a multi-region-based tumour sampling which, inherently, increases the costs of the experiments and may introduce - if not controlled - additional noise in the data due to the particular choices made in selecting the sampling sites. In order to control for these factors we propose to select the sampling regions based on their morphological characteristics, thus a pathologist could relatively easily identify them.

