ONCOBIOME: Gut OncoMicrobiome Signatures associated with cancer incidence, prognosis and prediction of treatment response

Funding: EU-H2020 (Grant agreement ID: 825410); Principal investigator for MUNI: Eva Budinska
Overview
(from the project’s official website)
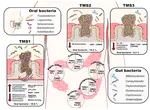
In view of the world-wide epidemic of cancer, which is linked to environmental and life-style factors, there is an unmet medical need in breakthrough concepts to understand the causes of neoplasia. One possible factor contributing to oncogenesis and tumor progression that has been neglected is the microbiota.
Indeed, the mammalian phenotype is largely driven by the combination of host and microbial genes. Beyond its role in regulating many fundamental functions of health, the intestinal metagenome appears to be dramatically implicated in cancer initiation, progression as well as responses to therapies, even for cancers developing in sterile tissues outside of the gastrointestinal tract.
Studies on the gut microbiome have so far been organized on a small and local scale, and there is an urgent need to:
- Fully identify and functionally characterize cancer-relevant microbial species by means of robust and standardized methods to validate cancer-associated gut microbiome fingerprints
- Assess their clinical relevance for patient prognosis
- Develop diagnostic tools that could become part of the oncological arsenal for the prevention, prediction and personalization of cancer therapy.
Project
Accumulating evidence underscores the role of the gut microbiome in health and disease, including cancer. The scope of the EU-funded ONCOBIOME project is to identify and functionally characterise the commensal bacteria of high clinical relevance. Using cohorts of more than 3 000 patients with cancer across 10 countries, project partners will identify microbiome signatures related to cancer occurrence, prognosis and response to therapy. They will also investigate the mechanistic interplay with the host metabolism, immunity and oncogenesis. Apart from fundamental knowledge, project results will lead to novel diagnostic and prognostic tests for cancer, as well as tailor-made pre- and pro-biotics.
Objectives
Beyond the role the intestinal metagenome plays in regulating multiple physBeyond its role in regulating multiple physiological functions that impact health, the intestinal metagenome is implicated in cancer initiation, progression and responses to therapies, even for extraintestinal neoplasia. Hence, there is an urgent need to fully identify and functionally characterize minimalist commensal ecosystems relevant to cancer, with reliable and robust methods, to validate cancer-associated gut microbiome fingerprints of high clinical relevance, and to develop diagnosis tools that will become part of the oncological arsenal for the optimization and personalization of therapy. Based on retro-and pro-spective studies, with large discovery and validation cohorts enrolling >9,000 cancer patients across 10 countries, ancillary to ongoing innovative clinical trials or FDA/EMA approvals across 4 frequent cancer types, ONCOBIOME will pursue the following aims: 1/ identify and validate core or cancer-specific Gut OncoMicrobiome Signatures (GOMS) associated with cancer occurrence, prognosis, response to, or progression on, therapy (polychemotherapy, immune checkpoint inhibitors, dendritic cell vaccines) or adverse effects, 2/ decipher the functional relevance of these cancer-associated gut commensal ecosystems in the regulation of host metabolism, immunity and oncogenesis, 3/ integrate these GOMS with other oncology hallmarks (clinics, genomics, immunomics, metabolomics) 4/ design optimal companion tests, based on those integrated signatures to predict cancer occurrence and progression. With high carat interdisciplinary experts, ONCOBIOME expects to validate cancer or therapy-specific Gut OncoMicrobiome Signatures (GOMS) across breast, colorectal, melanoma and lung cancers adjusting for covariates, to unravel the mode of action of these GOMS in innovative platforms, thus lending support to the design of cancer preventive campaigns using well characterized pre-and pro-biotics.